-Search query
-Search result
Showing 1 - 50 of 4,910 items for (author: shi & p)

EMDB-44372: 
In-cell Saccharomyces cerevisiae nuclear pore complex with single nuclear ring
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-44377: 
In-cell Saccharomyces cerevisiae nuclear pore complex with double nuclear ring and basket
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-44379: 
In-cell Mus musculus nuclear pore complex with nuclear basket
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-44381: 
In-cell Toxoplasma gondii nuclear pore complex
Method: subtomogram averaging / : Singh D, Hutchings J, Li Z, Guo Q, Villa E

EMDB-45197: 
In-cell Saccharomyces cerevisiae symmetry-expanded nuclear pore complex with double nuclear ring and basket
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45198: 
In-cell Saccharomyces cerevisiae symmetry-expanded nuclear pore complex with single nuclear ring
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45199: 
In-cell Saccharomyces cerevisiae nuclear pore complex cytoplasmic ring focused refinement
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45200: 
In-cell Saccharomyces cerevisiae nuclear pore complex inner ring focused refinement
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45201: 
In-cell Saccharomyces cerevisiae nuclear pore complex single nuclear ring focused refinement
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45202: 
In-cell Saccharomyces cerevisiae nuclear pore complex double nuclear ring focused refinement
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45203: 
In-cell Saccharomyces cerevisiae nuclear pore complex nuclear basket focused refinement
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45204: 
In-cell Saccharomyces cerevisiae nuclear pore complex membrane focused refinement for single nuclear ring
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45205: 
In-cell Saccharomyces cerevisiae nuclear pore complex membrane focused refinement for double nuclear ring
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45216: 
In-cell Mus musculus nuclear pore complex with nuclear basket consensus map
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45219: 
In-cell Mus musculus nuclear pore complex cytoplasmic ring focused refinement
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45220: 
In-cell Mus musculus nuclear pore complex inner ring focused refinement
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45222: 
In-cell Mus musculus nuclear pore complex nuclear ring focused refinement
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45223: 
In-cell Mus musculus nuclear pore complex basket focused refinement
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45227: 
In-cell Mus musculus nuclear pore complex membrane focused refinement
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45228: 
In-cell Toxoplasma gondii symmetry-expanded nuclear pore complex
Method: subtomogram averaging / : Singh D, Hutchings J, Li Z, Guo Q, Villa E
EMDB-45255: 
In-cell Saccharomyces cerevisiae C8-symmetrised nuclear pore complex consensus map
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45256: 
In-cell Saccharomyces cerevisiae symmetry-expanded nuclear pore complex consensus map
Method: subtomogram averaging / : Singh D, Hutchings J, Villa E

EMDB-45257: 
In-cell Mus musculus nuclear pore complex with nuclear basket consensus map
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45258: 
In-cell Mus musculus symmetry-expanded nuclear pore complex with nuclear basket consensus map
Method: subtomogram averaging / : Hutchings J, Singh D, Villa E

EMDB-45259: 
In-cell Toxoplasma gondii C8-symmetrised nuclear pore complex consensus map
Method: subtomogram averaging / : Singh D, Hutchings J, Li Z, Guo Q, Villa E

EMDB-42487: 
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer of dimers
Method: single particle / : Oldham ML, Qayyum MZ

EMDB-45005: 
Paired Helical Filament of tau amyloids found in Down Syndrome individuals
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D

EMDB-45007: 
Straight Filament of tau amyloids found in Down Syndrome individuals
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D

EMDB-45008: 
Paired Helical Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D

EMDB-45009: 
Straight Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D
PDB-9bxi: 
Paired Helical Filament of tau amyloids found in Down Syndrome individuals
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D
PDB-9bxo: 
Straight Filament of tau amyloids found in Down Syndrome individuals
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D
PDB-9bxq: 
Paired Helical Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D

PDB-9bxr: 
Straight Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids
Method: helical / : Tse E, Ghosh U, Condello C, Southworth D
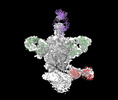
EMDB-44246: 
Cryo-EM structure of HIV-1 JRFL v6 Env in complex with vaccine-elicited, Membrane Proximal External Region (MPER) directed antibody DH1317.4.
Method: single particle / : Acharya P, Parsons R, Janowska K, Williams WB, Alam M, Haynes BF

EMDB-43128: 
Structure of the human dopamine transporter in complex with beta-CFT, MRS7292 and divalent zinc
Method: single particle / : Srivastava DK, Gouaux E
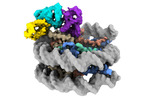
EMDB-38099: 
Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy
Method: single particle / : Ai HS, Tong ZB, Deng ZH, Pan M, Liu L
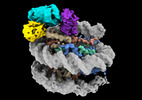
EMDB-38100: 
Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy
Method: single particle / : Ai HS, Tong ZB, Deng ZH, Pan M, Liu L
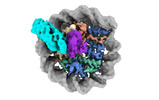
EMDB-38101: 
Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination)
Method: single particle / : Ai HS, Tong ZB, Deng ZH, Pan M, Liu L
EMDB-38102: 
Cryo-EM map of RNF168/UbcH5c-Ub/nucleosome determined by E2-Ub-NCP conjugation strategy
Method: single particle / : Ai H, Zebin T, Deng Z, Pan M, Liu L

EMDB-36914: 
Cryo-EM structure of Streptomyces coelicolor transcription initiation complex with the global transcription factor AfsR
Method: single particle / : Lin W, Shi J, Xu JC
EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)
Method: single particle / : Banerjee R, Khanppnavar B, Maharana J, Saha S, Korkhov VM, Shukla AK

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)
Method: single particle / : Banerjee R, Khanppnavar B, Maharana J, Saha S, Korkhov VM, Shukla AK
EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)
Method: single particle / : Banerjee R, Khanppnavar B, Maharana J, Saha S, Korkhov VM, Shukla AK

PDB-8jps: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)
Method: single particle / : Banerjee R, Khanppnavar B, Maharana J, Saha S, Korkhov VM, Shukla AK

EMDB-43746: 
Plasmodium falciparum 20S proteasome bound to an inhibitor
Method: single particle / : Han Y, Deng X, Ray S, Chen Z, Phillips M

PDB-8w2f: 
Plasmodium falciparum 20S proteasome bound to an inhibitor
Method: single particle / : Han Y, Deng X, Ray S, Chen Z, Phillips M

EMDB-42400: 
RORC mRNA 3'UTR riboswitch A97G/G98A mutant class C
Method: single particle / : Asarnow D, Khoroshkin M, Goodarzi H, Cheng Y

EMDB-42401: 
RORC mRNA 3'UTR riboswitch 77-GA mutant class A
Method: single particle / : Asarnow D, Khoroshkin M, Goodarzi H, Cheng Y

EMDB-42403: 
RORC mRNA 3'UTR riboswitch 117-AC mutant class C
Method: single particle / : Asarnow D, Khoroshkin M, Goodarzi H, Cheng Y
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
